Profiling with Sun Studio Analyzer
- Details Hits: 12528
This tutorial is based on fep.grid.pub.ro, the NCIT-Cluster Front End Processor. Everything you want to know about Sun Studio Analyzer is here.
Nobody says it better then Sun: watch this video tutorial about Sun Studio Analyzer and check the links section.
First, you need X Windows System forwarding. If you use Windows click here. and install Xming X Server for Windows.
Linux:
ssh This email address is being protected from spambots. You need JavaScript enabled to view it. -X
I. Start Sun Studio Analyzer:
[andrei.dumitru@fep ~]$ analyzer
II. Collect Experiment data
1. Go to File -> Collect Experiment...
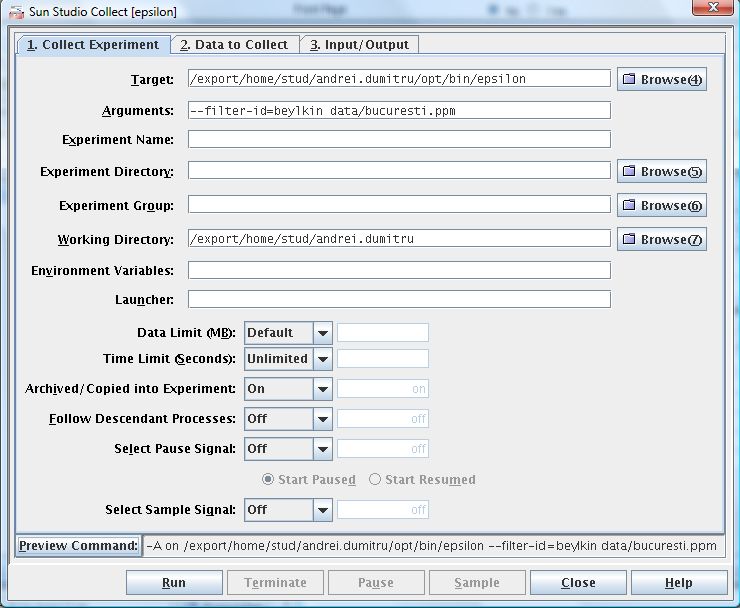
2. Select the Target , Working Directory and add Arguments if you need to.
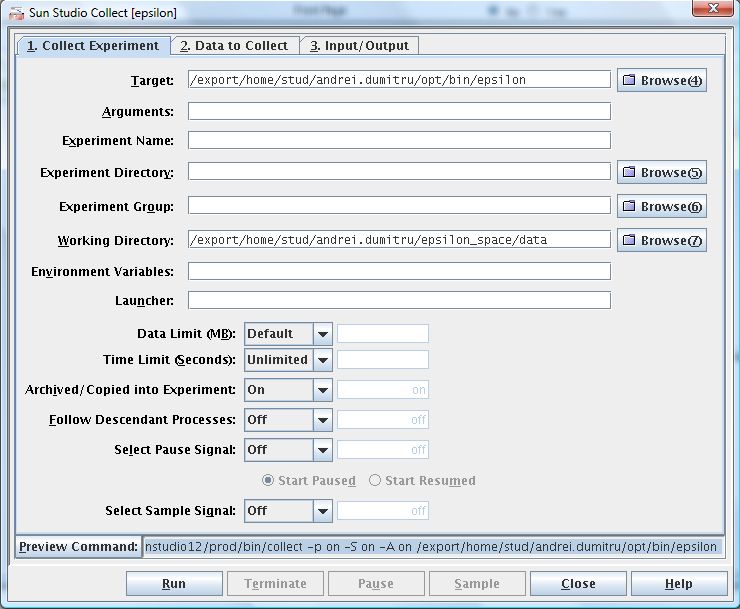
3. Click on Preview Command to view the command for collecting experiment data only.
4. Run your job on the cluster and wait for the results
[andrei.dumitru@fep ~]$ qsub -q ibm-quad -pe ibm-quad 2 -cwd -b y \
"/opt/openmpi/gnu-gcc/bin/mpirun -np 8 /opt/sun/sunstudio12/prod/bin/collect \
-p on -m on -S on -A on /export/home/stud/andrei.dumitru/opt/bin/epsilon \
--filter-id=beylkin data/bucuresti.ppm"
Highlighted with red is the collect command. In blue you can see the mpirun command and with pink the command to submit jobs
to the ibm-quad queue.
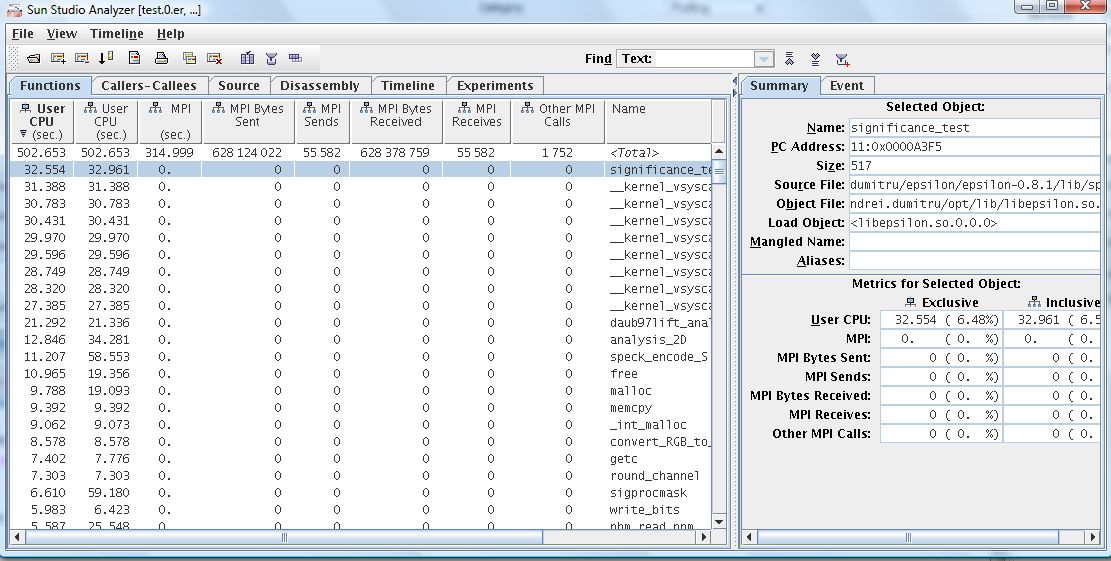
5. Open the results
Go to File -> Open Experiment...
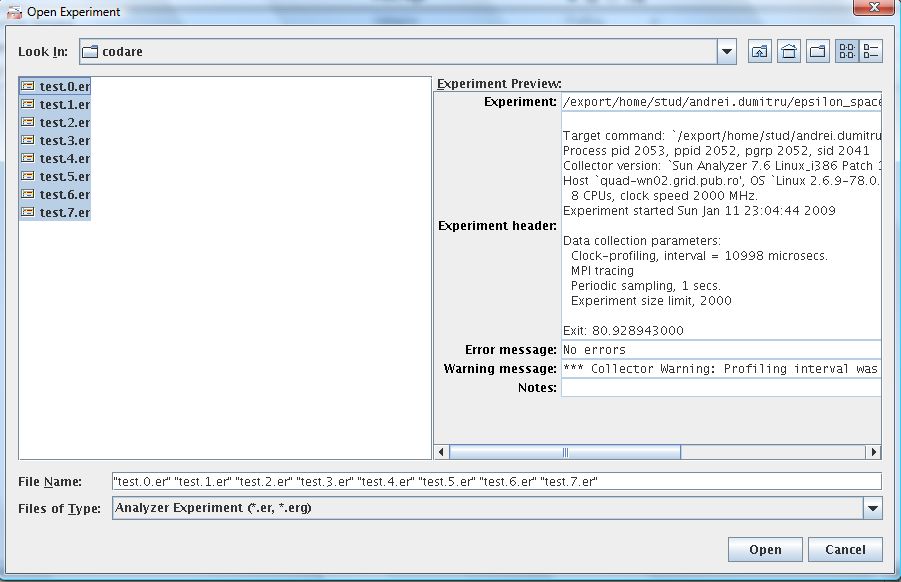
7. Select all the experiments you want to open.
8. Enjoy...